Simulation
CellDesigner is a modeling tool of biochemical networks with graphical user interface. Supporting SBML (Systems Biology Markup Language) format and SBW (Systems Biology Workbench) compliant, you can conduct simulation seamlessly via Simulation menu as well as via SBW.
CellDesigner can be used as a kind of SBML file editors for simulators.
There are two ways to conduct the simulation by CellDesigner:
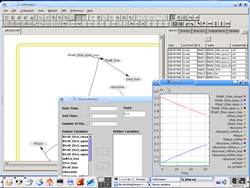
from SBW menu: to call SBML compliant simulators. |
|
 |
 |
Jarnac, Gibson simulator, etc... |
- Using the sample "simulation.html" - (StartUp Guide: chap.10 Simulation) (pdf)
- how to connect SBW (Help)
- Set the Kinetic Laws / Parameters
- Set the values of the Species
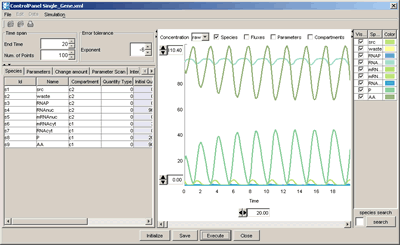
- Using Control Panel for simulation (Help)
Simulating SBML files
Examples
- Hypothetical single-gene oscillatory circuit in a eukaryotic cell via Simulation menu | (via SBW)
- Minimal Mitotic Oscillator via Simulation menu | (via SBW)
Working with Jarnac via SBW menu |
Working with Gibson Simulator on Linux |
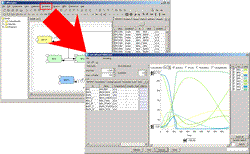
Simulation Results
CellDesigner open the Models from BioModels.net directly from [Database] menu.