Model to simulate
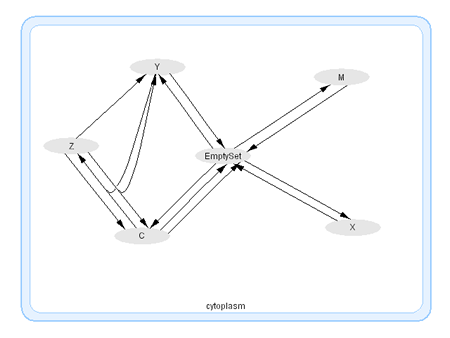
This is a modification of the widely cited Goldbeter (1991) minimal (3-variable) model for a mitotic oscillator. The three variables represent Cyclin ( C), inactive cdc-2 Kinase (M), and an active cdc-2 Kinase (X). Two additional variables Y, Z control the dynamics of the inhibitor.
Reference:
Gardner TS, Dolnik M, Collins JJ. A theory for controlling cell cycle dynamics using a reversibly binding inhibitor. PNAS 95: 14190-14195 (1998).


