Simulation Example 1:
Hypothetical single-gene oscillatory circuit in a eukaryotic cell
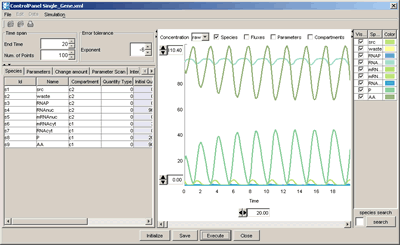
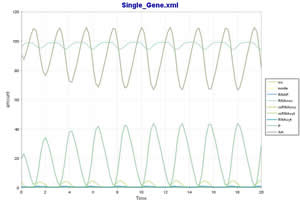
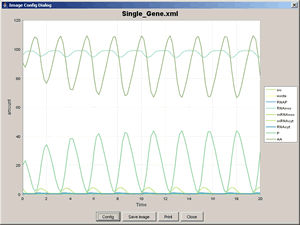
Here we try to simulate the model of a hypothetical single-gene oscillatory circuit in a eukaryotic cell using the SBML ODE Solver directly invoked via CellDesigner's Simulation menu. This model was proposed by Michael Hucka, Andrew Finney, Herbert Sauro, Hamid Bolouri, et al. in [1]. Please refer the paper for more detailed information.
Model to simulate
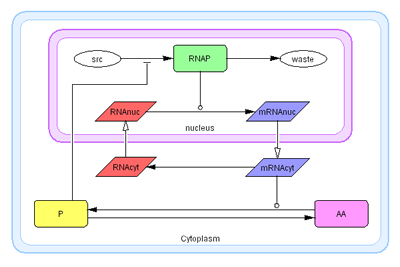
In this model, the nucleus of the cell is represented as one compartment
and the surrounding cell cytoplasm as another compartment.
- src (source material)
- waste (degradation)
- RNAP (RNA Polymelase)
- RNAnuc (RNA in nucleus)
- RNAcyt (RNA in cytoplasm)
- mRNAnuc (mRNA in nucleus)
- mRNAcyt (mRNA in cytoplasm)
- P (product)
- AA (amino acids)
 Model file"Single_Gene.xml" to download
Model file"Single_Gene.xml" to download