CellDesigner News
CellDesigner on Garuda platform (2017/2/14) Garuda enabled Ver4.4 is available as Ver4.4.1 Win / Mac.
PhysioDesigner 1.0 is available. PhysioDesigner can embed CellDesigner's SBML model into its multilevel physiological PHML model. (2014/08/01)
CellDesigner 4.4 is now available (2014/07/12)
Plugin: iPathwaysPlus Register. Check Plugins page...(2014/01/27)
Model Repository is updated (2013/Oct)
CellDesigner 4.3 is now available (2013/02/01)
PhysioDesigner 1.0beta is available. (2011/06/22)
Plugin: BioPAXLevel 3 Export plugin is now available (2011/11/8)
CellDesigner 4.2 is now available (2011/10/05)
Ver.4.2 supports Reduced Notation, Simulation Parameter Polymorphism, SBMLsim solver... and more versionup info...
Tool: iPathways 1.2 (IAppli) released (2011/08/30)
You can access the colleection of the pathway models from your iPod/iPad! Check iPathways.org!
Plugin: SBSI CellDesigner plugin. (2011/01/19)
Check Plugins page...
Plugins: Merge Models & Payao Uploader. (2010/10/10)
Check Plugins page...
Plugin: PathwayAccess: CellDesigner Plugins for Pathway Databases (2010/07/28)
Check Plugins page...
New Model using CellDesigner (2010/07/28)
CellDesigner 4.1 is now available (2010/06/28)
Ver.4.1 is SBML Level2 Version4 Support, BioPAX Level 3 export and SABIO-RK integration..... and more versionup info...
A Comprehensive Molecular Interaction Map for Rheumatoid Arthritis. 2010, PLoS ONE uses CellDesigner (2010/04/26)
CellDesigner 4 Extension Tag Specification Document is available. (2010/04/04)
SBMLsqueezer Ver1.3 is released! (2010/04/04)
It can run as a CellDesigner plugin (2010/04/04)
CellPublisher is available. (2010/04/04)
CellPublisher is a free and open web server to share CellDesigner diagrams with a wide audience.
Sample plugin for CellDesigner 4.1beta: (2009/10/16)
MappyingArrayMass readme and sample data is now available
Winter School on Computational Modeling in Biology
December 14-18, 2009, Okinawa Institute of Science and Technology (OIST). Data and model sharing by SBML and insilicoML. For details: http://www.mei.osaka-u.ac.jp/gCOE/school/2009_winter/
CellDesigenr Tutorial at ICSB-2009 (Aug 30th 12:15-14:45)
For Mac Leopard (10.5.7) Users (July 2008)
Due to the recent Mac Java update, there is a problem of CellDesigner launch. The fix patch is now available here.
Import from BioModels.net
Recent models in BioModels.net are in SBML Level 2 Version 3 format. Current CellDesigner 4.0.1 can import up to SBML Level 2 Version 1. Please wait for the coming release of CellDesigner to import the SBML Level 2 version 3 models. >BioModels.net models simulation results
CellDesigner Tutorial at ICSB-2008(Aug 22nd, 2008)
CellDesigner Tutorial Presentation file is available here (in PDF format) .
Panther Pathway released a new Viewer Applet compatible with CellDesigner 4.0 (Aug 21, 2008)
PANTHERTM Pathway (http://www.pantherdb.org/pathway/) released a new version of the PANTHER Pathway Viewer Applet, which is compatible with the newly released CellDesigner 4.0 and Systems Biology Graphical Notation (SBGN) Process Diagram Level 1 (http://www.sbgn.org/). It is a result of the collaboration between by the Evolutionary Systems Biology Group (http://www.ai.sri.com/esb/) at SRI International and SBI (http://sbi.jp), and the work is funded by NEDO and NIH.
PANTHERTM Pathway is one of the modules within the PANTHERTM Classification System (http://www.pantherdb.org/). The current release contains 165 signaling and metabolic pathways created using CellDesigner and curated using the PANTHER Pathway Curation System (http://curation.pantherdb.org/). The PANTHER Pathway Curation System is available for public use.
CellDesigner 4.0.1 Released!(Aug 12nd, 2008)
CellDesigner 4.0 Released! (August 4, 2008)
Encounter Install Error? check libSBML version.. (June 30, 2008)
Plugins / Utilities page is now available (May 1, 2008)
CellDesigner is featured in front page of Science web site. (April 29th, 2008)
A Comprehensive map of RB/E2F pathway (using CellDesigner v3.0) published online in Molecular Systems Biology (Mar 6th, 2008)
BioModels.net Simulaton Results (Dec 20th, 2007)
BioModels.net Simulaton Results with CellDesigner 3.5.2 is now available.
CellDesigner 4.0 beta Released! (Dec 7th, 2007)
Ver.4.0 beta is the Graphical Notation Preview version, preview for the new graphical notation conform to SBGN Level 1 draft..... and more versionup info...
CellDesigner download tops 22,000! (Dec 1st, 2007)
THANK YOU for using CellDesigner! Your feedback is always welcome!
CellDesigner 3.5.2 Released! (Nov 15th, 2007)
Bug Fix for Ver.3.5.1 ...
CellDesigner Tutorial at ICSB-2007 (Oct 3rd, 2007)
CellDesigner Tutorial at ICSB-2007 Presentation file now available (Oct 3rd, 2007)
CellDesigner Tutorial Presentation file is available here (in PDF format 5.8MB) .
CellDesigner Tutorial at ICSB-2007 (Aug 15th, 2007)
CellDesigner Tutorial at ICSB-2007 Tutorial Registrataion is now open at ICSB-2007 website.
Call for Participation on Pathway Curation (Mar 1st, 2007)
PANTHER Pathway is a community effort to build up the pathway database with curation. We give full credit to all our contributors on PANTHER website. Please contact pathcuration at pantherdb.org for any questions.
CellDesigner 3.5.1 Released! (Jan 10th, 2007)
Bug Fix for Ver.3.5 ...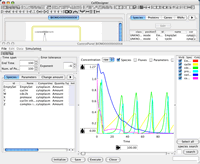
CellDesigner 3.5 Released! (Dec 28th, 2006)
Ver.3.5 now supports smooth SBML file handling. Validate and auto-layout when opening a SBML file, simulate and output to PDF for your publication... and more versionup info...
CellDesigner download tops 15,000! (Dec 26th, 2006)
THANK YOU for using CellDesigner! Your feedback is always welcome!
Send us your comments on CellDesigner! (Nov 8th, 2006)
Please answer to CellDesigner survey for your feedback / feature requests for CellDesigner.
CellDesigner Tutorial at ICSB-2006 Presentation file now available (Oct 9th, 2006)
 CellDesigner Tutorial Presentation file is available here (PDF 5.8MB) .
CellDesigner Tutorial Presentation file is available here (PDF 5.8MB) .
CellDesigner 4.0 alpha Released! (Oct 6th, 2006)
Ver.4.0 alpha is the Plugin Developer's version, preview for the new feature which enables development plugin modules onto CellDesigner.... and more versionup info...
CellDesigner Tutorial at ICSB-2006 (Aug 11th, 2006)
 CellDesigner Tutorial is planned to be held on October 8th, 2006 at Pacifico Yokohama Conference Center as part of ICSB-2006 tutorial programs.
CellDesigner Tutorial is planned to be held on October 8th, 2006 at Pacifico Yokohama Conference Center as part of ICSB-2006 tutorial programs.
CellDesigner 3.2 Released! (July 5th, 2006)
Ver.3.2 now support libSBML, Java5 and IntelMac. Direct connect to Entrez Gene, output to PDF... and more versionup info...
CellDesigner 3.1 Released! (2006.3.24)
CellDesigner3.1aims to achieve smoother, user friendly operation such as performance tuning, import models from BioModels.net...
versionup info...
CellDesigner download tops 10,000! (2006.3.7)
THANK YOU for using CellDesigner! Your feedback is always welcome!
For Mac OS 10.3.9 User (2006.1.27)
There is a Mac OS X bug that affects CellDesigner on OS X 10.3.9 systems with the QuickTime 7.0.4 upgrade. This appears to only affects systems running 10.3.9, and not 10.4. The current workaround is to revert to QuickTime 7.0.1, through a reinstaller available from Apple on the Support/Downloads page.
c.f. - InstallAnywhere Support information
CellDesigner 3.0.1 Released (2006.1.26)
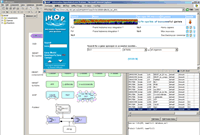 CellDesigner3.0.1 fixed bugs on complex handling..
CellDesigner3.0.1 fixed bugs on complex handling..
versionup info...
CellDesigner 3.0 Released (2005.10.15)
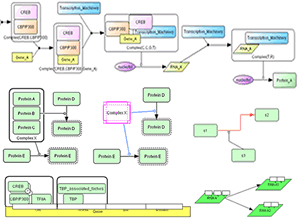 CellDesigner 3.0 now supports new graphical notation as well as direct simulation using the intuitive control panel.
CellDesigner 3.0 now supports new graphical notation as well as direct simulation using the intuitive control panel.
versionup info...
CellDesigner 2.5 Released (2005.9.1)
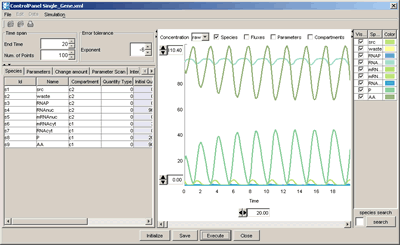 CellDesigner 2.5 now support direct simulation using the intuitive control panel.
CellDesigner 2.5 now support direct simulation using the intuitive control panel.
versionup info...
CellDesigner 3.0 Alpha Notation Preview Version Released (2005.8.4)
CellDesigner 3.0 alpha version is to preview the new notation scheme.
versionup info...
CellDesigner 2.2 Released (2005.5.24)
CellDesigner now supports DB connections and SVG export.
Referring to databases such as DBGET and PubMed is now available.
You can also export image to SVG format.
versionup info...
CellDesigner 2.1 Released (2005.1.12)
CellDesigner now supports newly released SBW2.0.
Version 2.0 Download Tops 1000 in three months (2005.1.12)
More than 1000 copies of CellDesigner 2.0 have been downloaded since October, 2004.
PANTHER Pathway Module released (January 3, 2005)
 Applied Biosystems Inc. (ABI) today released PANTHERTM pathway module that contains over 100 signal transduction pathways all created using CellDesigner. It is a results of a year collaboration with ABI and the Systems Biology Institute (SBI) to represent molecular interactions involved in signal transduction pathways with rich information contents and access to genomic databases. .....
Applied Biosystems Inc. (ABI) today released PANTHERTM pathway module that contains over 100 signal transduction pathways all created using CellDesigner. It is a results of a year collaboration with ABI and the Systems Biology Institute (SBI) to represent molecular interactions involved in signal transduction pathways with rich information contents and access to genomic databases. .....
(ABI press release)
> Related Paper : Huaiyu Mi, et al
The PANTHER database of protein families, subfamilies, functions and pathways.
Nucl. Acids. Res., 31:334-341. 2005
Diabetes-Related Molecular Interaction Map Released (2004.12.01)
Molecular interaction maps for adipocyte, pacreatic beta-cell, skeletal muscle cell, and hepatocyte are released as a part of paper published in Diabetes journal (paper URL:http://www.symbio.jst.go.jp/symbio2/papers/Diabetes2004.pdf). The map covers type-II diabetes related molecular interactions for four cell types that are heavily interrelated.
Each map can be access from the SBI Model Repository (URL:http://systems-biology.org/001/003.html).
Macrophage Molecular Interaction Map using CellDesigner Published (2004.09.30)
A team of researchers at the Systems Biology Institute (SBI), Tokyo Medical and Dental University (TMDU), and Keio University jointly created a large scale molecular interaction map of macrophage. The map is intended to serve analysis framework for the Alliance for Cellular Signaling (AfCS), a large scale NIH-funded project lead by Professor Alfred Gilman (Univ. Texas Southwestern Medical Center). The map and description of the map is available from Nature Signaling Gateway (http://www.signaling-gateway.org/reports/v2/DA0014/MacrophageMolecularInteractionMapVer1.0.pdf).
This map was created with CellDesigner version 2.0 (http://www.systems-biology.org/002/). A total number of 506 reactions and 678 species were included. Most signal transduction pathways that are used in ligand screening experiments are included, as well as basic metabolic circuits and secration pathways.
CellDesigner 2.0 Released (2004.9.30)
CellDesigner 2.0 was released on September 30, 2004. A new version features SBML Level 2 support, enhanced graphical notation system (with new symbols and color representation), intuitive user interface and information organizing functionalities, block diagram editor to describe the modifications/activations logics.
CellDesigner downloads Top 2500 (2004.07.12)
More than 2500 copies of CellDesigner has been downloaded in a year since its release in June 2003. More than 1000 registrants are using CellDesigner today.

