CellDesigner 4.0
 CellDesigner 4.0 is the enhanced Graphical Notation version.
CellDesigner 4.0 is the enhanced Graphical Notation version.
* For upgrading users from CellDesigner ver.3.5.2 / 4.0beta, please check the File compatibilities.
 Major New Features:
Major New Features:
- Enhanced Graphical Notation
CellDesigner's Graphical Notation conforms to SBGN (Systems Biology Graphical Notation) proposal of April 2008 draft (* not complete SBGN Process Diagram Level1 which became available in August 2008).
- Enhanced KineticLaw Edit Dialog:
MathML rendering in KineticLaw.
- Simulation / Control Panel enhancement
- Export Species/Reactions information including Notes into CSV file
- Validation of files when opening (libSBML3)
- Native file chooser support for Mac OS X
- Enhanced Edit Functions
- [Layout] menu: Automatic Layout function (using yFiles layout library.)
- Layer Function (alpha version)
to add comments / arrows on top of the models.
- Database link to Genome Network Platform

- Plugin development framework.
Develop your plugin on Eclipse, and you can call the plugin from [Plugin] menu.
> Plug-In Tutorial [pdf] > http://celldesigner.org/plugins.html
- Export of Pure Level 1 Version 1 is no longer supported.
- Windows 2000 is no longer supported.
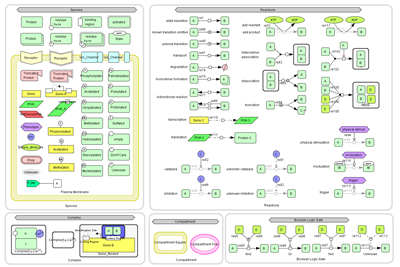
Graphical Notation:
CellDesigner 4.0 now enables you to preview for the new graphical notation conform to SBGN Level 1 draft.
(*click to enlarge the upper right image.)
(* not complete SBGN Process Diagram Level1 which became available in August 2008).
 New Species: Drug, Tag
New Species: Drug, Tag - New Reactions: physical stimulation, modulation, trigger, Boolean logic gates
- Process nodes of the reactions are introduced to connect modifiers such as catalysis, inhibition.
- Protein states (open, close), binding region

- Protein residue modification types enhanced (Glycosylated, Myristoylated, Palmtoylated, Prenylated, Protonated, Sulfated)
- Gene modification regions enhanced: (phospholyrated, acetylated, methylated),
coding regions, regulatory regions, transcriptionstartingSiteL/R
- Allow to draw Complexes within a Complex
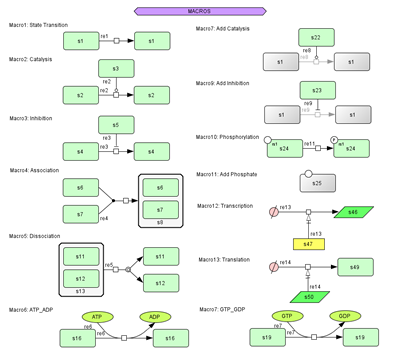
Macro Function:
- 13 quick-to-draw sets of components available.
KineticLaw Editor: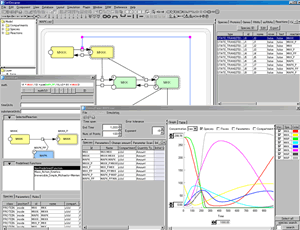
- Preview for the selected reaction in the editor
- Utilize Predefined Function for "math"
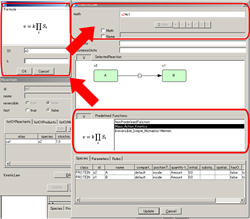
MathML rendering in "math"
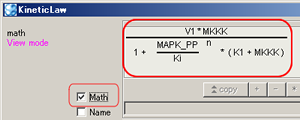 |
Switch ID/Name display in the "math"
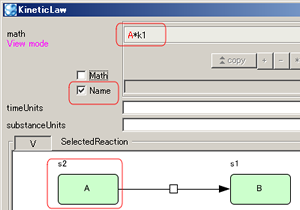 |
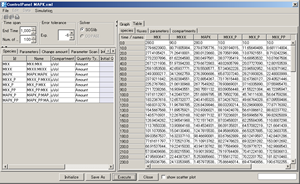 Control Panel:
Control Panel:
- Copasi integration >> How to setup Copasi
- Solver selection in the ControlPanel : SOSlib and Copasi
- Simulation results displayed in Table format
- Show Scatter Plot
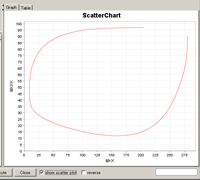
File:
- Export Species / Reactions information including Notes into CSV file.
- Validation of files when opening (libSBML3)
- Native file chooser support for Mac OS X
Edit:
- Changes color/shape change for multiple Species/Reactions.
- Highlight List of Proteins selected in the Draw area.
- Sort (ascend/descent) by click in the list
- [Find All] menu added.
- [Species ID Replace] dialog
- Edit the position of the name of the Compartment display
- Reaction ID Display on screen
- Toolbar display ON/OFF by "View-Change Toolbar Visible" menu
User Interface Enhancements:
- Change display position of the list: [View]-[List] menu, then [Right] or [Down]
- Display Reaction ID on screen: [View]-[Show Reaction ID]
- Change display position of Compartment name: drag the name to the new position.
- Change Icon display size: [Preference] - [Set Icon Size]
- [Preference]-[Set Macro UI]
- Popup setting dialog: [Preference]-[Popup]
Layer Function (previsionary)
- to add comments / arrows on top of the models.
Installation:
New Installer is being adopted. For details on installation procedure, check here.
 Major New Features:
Major New Features:
 New Species: Drug, Tag
New Species: Drug, Tag 




 Control Panel:
Control Panel: 



