The current release is distributed in archived installer package for each operating system.
- Windows: CDInstall_2_1_win.exe, Setup_SBW_1_modules.exe (*)
- MacOSX: CDInstall_2_1_mac.zip
- Linux: CDInstall_2_1_linux.bin
- (*) Setup_SBW_1_modules.exe is to setup the SBW modules that CellDesigner uses.
While J2RE is required for CellDesigner to run, the installers for Windows and Linux include it, and MacOSX has a Java runtime environment initially. Therefore, you do not need to download or install J2RE. (If you installed SBW, you have already had it anyway.) Download the one for your operating system and follow instructions below to install CellDesigner for each environment.
Windows:
- Double click CDInstall_2_1_win.exe.
The installer window should open, and follow the message therein. - Select the install set according to your choice of SBW.
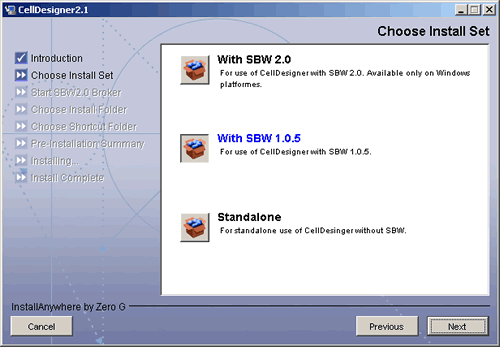
- *For those who have installed SBW 2.0,
when the message “Start SBW 2.0 Broker” is displayed,
Start “C++ Broker” by selecting Windows’ [Start] menu then – [Systems Biology Workbench] – [C++ Broker].
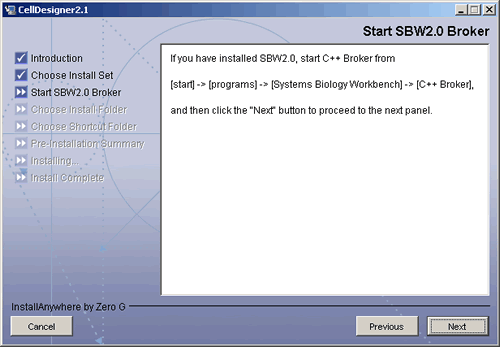
- Press [Next] to execute the rest of the installation.
- When completing the installation of CellDesigner, start SBW_1_modules.exe to install the SBW simulation modules.
Note: In case you have installed SBW 2.0 and you encounter an error while installing CellDesigner, there might be a possibility that the C++ Broker is up which prevents CellDesigner to start.Please try to kill the broker using the Task Manager, or restart your system before you resume the CellDesigner installation.
MacOSX:
- Double click CDInstall_2_1_mac.zip.
- The compressed installer should be recognized by Stuffit Expander and should automatically be expanded to CDIinstall_2_1. Then double click it.
The installer window should open, and follow the message therein.
Linux:
- Open a shell and, cd to the directory where you downloaded the installer.
- At the prompt type: sh ./CDInstall_2_0_linux.bin.
The installer window should open, and follow the message therein. - If you have installed SBW,
- Select the install set according to your choice of SBW.
- Set the path to the SBW library.
After then, an installer window should open, and follow the message therein.
If you have installed SBW, set the path to the SBW library.
[Installed File Images]
After installation finished, you would see the following directories/files in the installation directory (CellDesigner2.1 by default).
+00README.txt
|
+CellDesigner2.1 executable application module(*for Mac: CellDesigner)
+CellDesigner_register_to_SBW
|
+documents/
| +startup_guide21.pdfthis document
|
+exec/
| +celldesigner.jar library for CellDesigner application
|
+jre (1.4.2_06) *(Windows, Linux only)
|
+lib
| +sbw/
| +SBWCore.jar
| +browserlauncher/
| +browserlauncher.jarlibrary web browser launcher by Eric Albert
| +mrjadapter/
| +MRJAdapter.jarlibrary for MacOSX feature by Steve Roy, Software Design
| +xerces/
| +xercesImpl.jar XML library by the Apache Software Foundation
| +xml-apis.jarXML-API library by the Apache Software Foundation
|
+licenses/
| +mrjadapter/
| +lgpl.txt
| +xerces/
| +LICENCE
| +LICENCE-DOM.html
| +LICENCE-SAX.html
|
+samples/
| +components.xml sample for various components
| +M-Phase.xml sample for model editing
| +M-Phase2.xml sample for model editing
| +simulation.xmlsample for simulation
| +test-level1.xmlsample for SBML level 1 document
| +test-level2.xmlsample for SBML level 2 document
|
+Uninstall_CellDesigneruninstaller module directory
SBW_1_modules (installed by Setup_SBW_1_modules.exe)
+jre (1.3.1_11)
|
+licences
| +lgpl.txt
|
+modules
| +plot.jar
| +SimDriver.jar
|
+Uninstall_SBW_1_modules